Approach
Genome-to-genome interactions
We are interested in indentifying genome-to-genome interactions between Medicago truncatula and Meloidogyne Hapla.
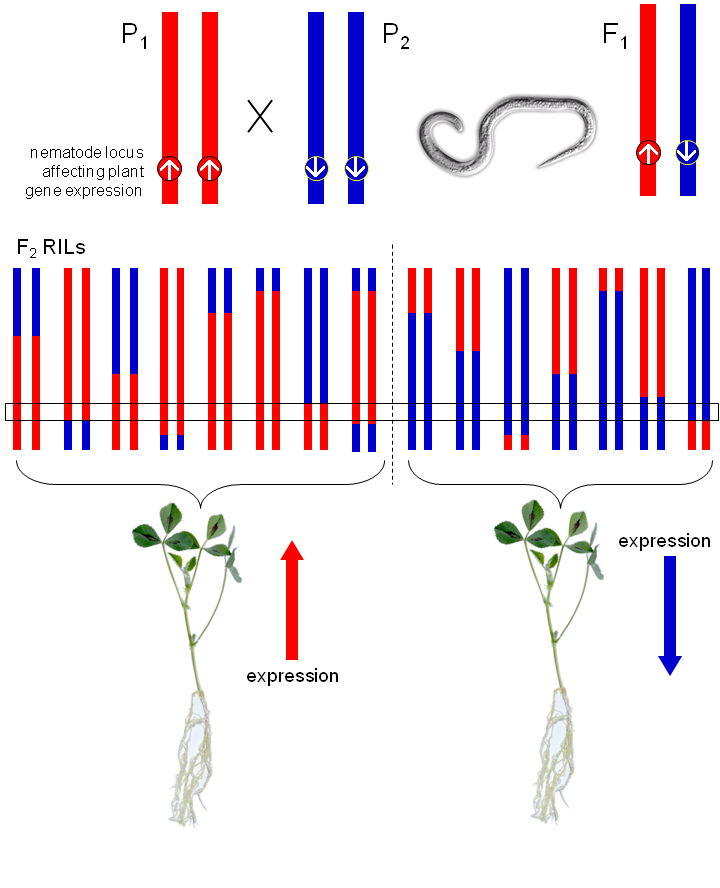
F2 recombinant inbred lines
The facultative sexual and asexual reproduction mechanisms of M. hapla allowed co-PI Valerie Williamson to generate an M. hapla mapping population composed of 183 F2 lines (Liu et al., 2007). This population was produced by crossing strain VW9 (the fully sequenced and annotated strain) and strain VW8. Due to a somewhat unusual meiosis, M. hapla selfed progeny are homozygous for most loci. This is caused by rejoining of sister nuclei of the second meiotic division in combination with apparent 4-strand crossovers (Liu et al., 2007). Because of this homozygosity, M. hapla F2 lines are genetically equivalent to recombinant inbred lines (RILs), and as such, are easily maintained.
Cross-species eQTL Mapping
We are mapping expression levels of the plant genes to genetic loci in the nematode. To accomplish this, we take advantage of the F2 RILs. Genetically homogeneous host plants are inoculated with a nematode line (one RIL per plant), and infected tissue is harvested for each plant at a fixed time point post-infection. These samples provide RNA from both species concurrently. RNA-seq is then used to assay the RNA. This provides gene transcript abundance quantification for both species, plus the ability to detect and genotype polymorphisms within the nematode lines. Gene expression levels in the plants can then be compared with the pathogen genotypes.