




Next: Bootstrapping
Up: Recreating Datasets
Previous: Recreating Datasets
Contents
Index
When you use the -b option with an integer value from one to six, Prune will
take one action, print out a new dataset and exit. There are six possible actions, and some require that
you use the -M option with a percentage as well. The six possible actions are listed below:
use the number preceeding the action with the -b option to do that action.
- Bootstrap the data. This option samples individuals with replacement to produce a new data set of the
same size as the original.
- Permute the data. This option will randomly shuffle the traits against the genotypic arrays. If there
are multiple traits, then there is a shuffling for each trait. If you want to keep trait arrays together, see
item six below.
- Simulate missing markers. This option requires you to specify a percent
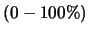 of missing marker information.
Prune will then randomly reset that percentage of marker data to unknown.
of missing marker information.
Prune will then randomly reset that percentage of marker data to unknown.
- Simulate dominant markers. Again, you need to specify the percentage of dominant marker systems.
A random set of markers are chosen and converted to dominant markers. The direction is random (50/50 each way)
for each marker chosen.
- Simulate selective genotyping. The -M option specifies what proportion of individuals that are typed.
Do this with single trait data sets. The individuals are ordered according to their trait values, and one-half
the specified proportion in the tails are retained in the output. The individuals in the middle of the
distribution are deleted from the data set.
- Permute the data. This is the same as option 2 above except that with multi-trait data sets, there is a shuffling
of the trait arrays against the genotype arrays. If you think the traits are correlated, you can use this to maintain
that correlation. A value of 12 is the same sas this option.





Next: Bootstrapping
Up: Recreating Datasets
Previous: Recreating Datasets
Contents
Index
Christopher Basten
2002-03-27