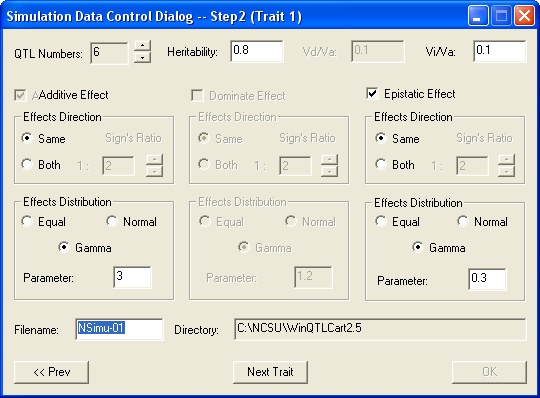
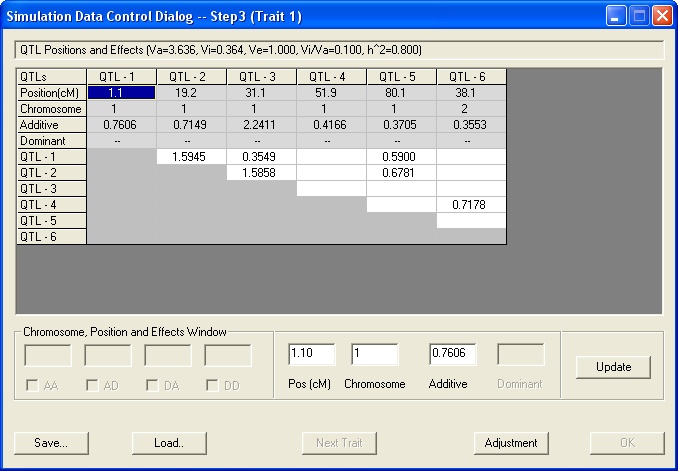
To create a data simulation file, select File>Simulation. This displays the Create Simulation Data series of dialogs.
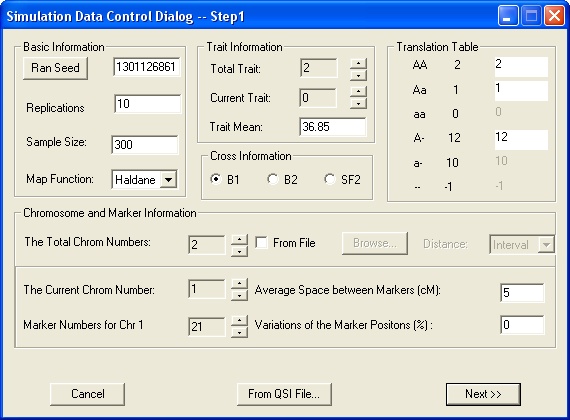
Fill in the blanks for yourself, or accept the defaults.
Sample size
Trait mean
Random seed. Click the button to generate a new random seed.
Map function. Haldane (assumes no crossover interference) or Kosambi (assumes some crossover interference).
Cross Information. Select an experimental design option. You can select only one option to modify at a time. (At this time, only the three options below are available for selection.)